Advanced Single-cell scATAC-seq & scRNA-seq Dual-omics Analysis: CopyscAT Analysis
Introduction
TIP
CopyscAT infers Copy Number Variations (CNV) based on scATAC-seq data, assisting in the identification of cancer cells. It helps investigate the relationship between chromosomal changes and epigenomic states of different subclones within complex tumors (such as glioblastoma), analyzing how genetic variations affect molecular phenotypes of cells.
In single-cell multi-omics research, scATAC-seq data not only provides information on chromatin accessibility but can also be used to infer Copy Number Variations (CNV) by analyzing read counts in genomic regions. CopyscAT uses single-cell epigenomic data to infer CNV, thereby defining and identifying cancer cells. The core principle of this tool is using the number of reads aligned to genomic regions in scATAC-seq data as a proxy metric for DNA copy number in those regions.
Core Functions of CopyscAT
Malignant Cell Identification Distinguishes malignant cells from non-malignant cells. A key advantage of CopyscAT is its ability to automatically identify non-tumor cells and use them as controls, thereby detecting CNV in tumor cells more accurately.
Multi-level CNV Detection Capable of detecting local CNV (such as ecDNA, i.e., extrachromosomal DNA), fragment-level CNV, and chromosome-arm-level CNV, comprehensively parsing genomic structural variations in tumors.
Tumor Heterogeneity Resolution Identifies subclonal populations with distinct CNV characteristics, particularly suitable for processing complex samples with high levels of intra-tumor heterogeneity, such as glioblastoma.
This document aims to provide a comprehensive technical guide for CopyscAT for single-cell multi-omics researchers, covering its basic principles, operation methods on the SeekSoul™ Online cloud platform, result interpretation, and frequently asked questions, helping you quickly master and apply this tool.
Theoretical Basis of CopyscAT
Core Principle
The core idea of CopyscAT analysis is: Using the number of reads aligned to genomic regions in scATAC-seq data as a proxy for DNA copy number in that region, inferring the copy number variation status of each cell through steps such as data normalization, CNV detection, and cell assignment.
What is CNV?
Copy Number Variation (CNV) refers to structural variations in the quantity of large DNA fragments in the genome, mainly manifesting as amplification (gain) or deletion (loss) of chromosomal regions. Under normal circumstances, human cells are diploid (typically having 2 copies of each autosome); when amplification occurs, the copy number exceeds 2, and when deletion occurs, the copy number is less than 2.
CNV is a key driver in the initiation and progression of many diseases, including tumors. Traditional CNV analysis is mainly based on Whole Genome Sequencing (WGS) or Whole Exome Sequencing (WES), which can only provide population-average information. Single-cell CNV analysis infers the copy number status of each cell at single-cell resolution, revealing genomic differences and heterogeneity among different cells within tissues.
Two Directions of Single-cell Multi-omics CNV Analysis
There are two main directions for using single-cell multi-omics data for CNV analysis:
- Direction 1: Based on scRNA-seq data - Infers CNV using gene expression information, with infercnv being the primary tool.
- Direction 2: Based on scATAC-seq data - Infers CNV using read counts, with primary tools including epiAneuFinder, AtaCNV, and CopyscAT.
TIP
Brief Introduction to Mainstream Single-cell ATAC-seq CNV Analysis Tools
Currently, tools commonly used for CNV detection in scATAC-seq data include: epiAneuFinder, CopyscAT, and AtaCNV. This guide focuses on the detailed usage of CopyscAT. If you need to use other tools, please refer to the relevant documentation for more information.
Key Algorithms and Workflow
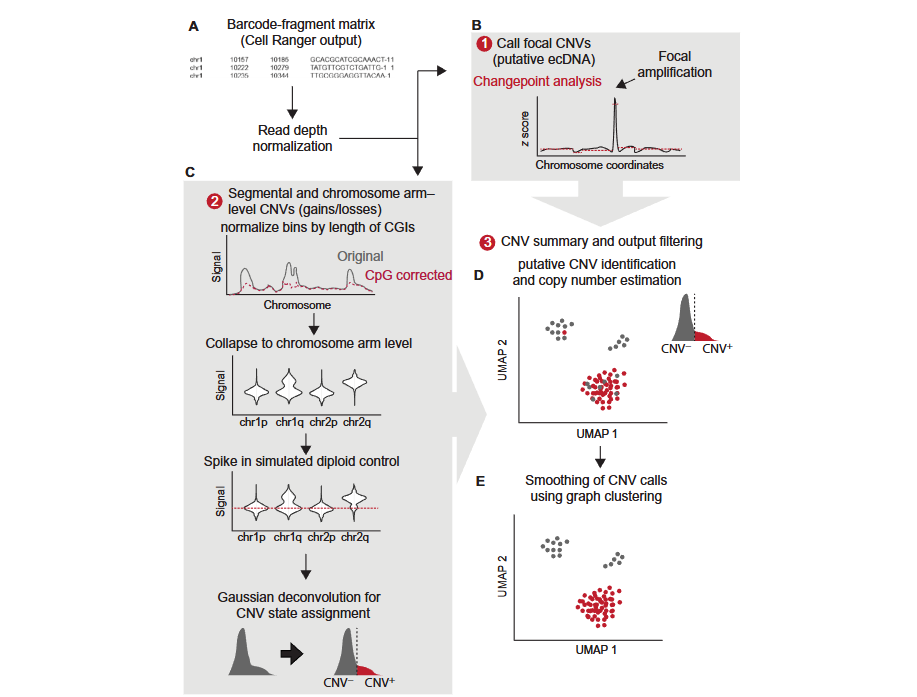
The analysis workflow of CopyscAT includes the following key steps:
- Data Input: Accepts the barcode-fragment matrix generated by seekARC as input.
- Data Normalization: Normalizes the coverage matrix to generate a normalized matrix.
- CNV Detection:
- Local CNV Detection: Infers local CNV (such as ecDNA, i.e., extrachromosomal DNA) through large peaks in the normalized coverage matrix.
- Fragment and Chromosome-arm Level CNV Detection: Uses the normalized matrix to infer fragment-level and chromosome-arm-level CNV.
- Double Minutes Detection: Identifies high copy number amplification regions in the genome.
- Cell Assignment: Uses consensus clustering methods to finally determine cell grouping and CNV status.
A key advantage of CopyscAT is its ability to automatically identify non-tumor cells and use them as controls, thereby detecting CNV in tumor cells more accurately. This tool is particularly suitable for processing complex samples with high levels of intra-tumor heterogeneity.
Applicable Scenarios and Main Objectives
Suitable Sample Types:
- Tumor Tissue Samples (Highly Recommended) - Contain abundant CNV events, revealing tumor heterogeneity and subclonal structures.
- Precancerous Lesions or Developmental Anomalies - Can detect early genomic structural variations.
Unsuitable Sample Types:
- Normal Healthy Tissues - Most cells are diploid and lack significant CNV events; CNV analysis has limited significance.
Main Objectives of Single-cell CNV Analysis:
- Malignant Cell Identification - Distinguishing malignant cells from non-malignant cells (malignant cells typically exhibit large-scale, non-random CNV patterns).
- Tumor Heterogeneity Resolution - Identifying subclonal populations with distinct CNV characteristics.
- Clonal Evolution Tracking - Inferring clonal evolutionary relationships of tumors through CNV pattern similarity.
Cloud Platform Operation Guide
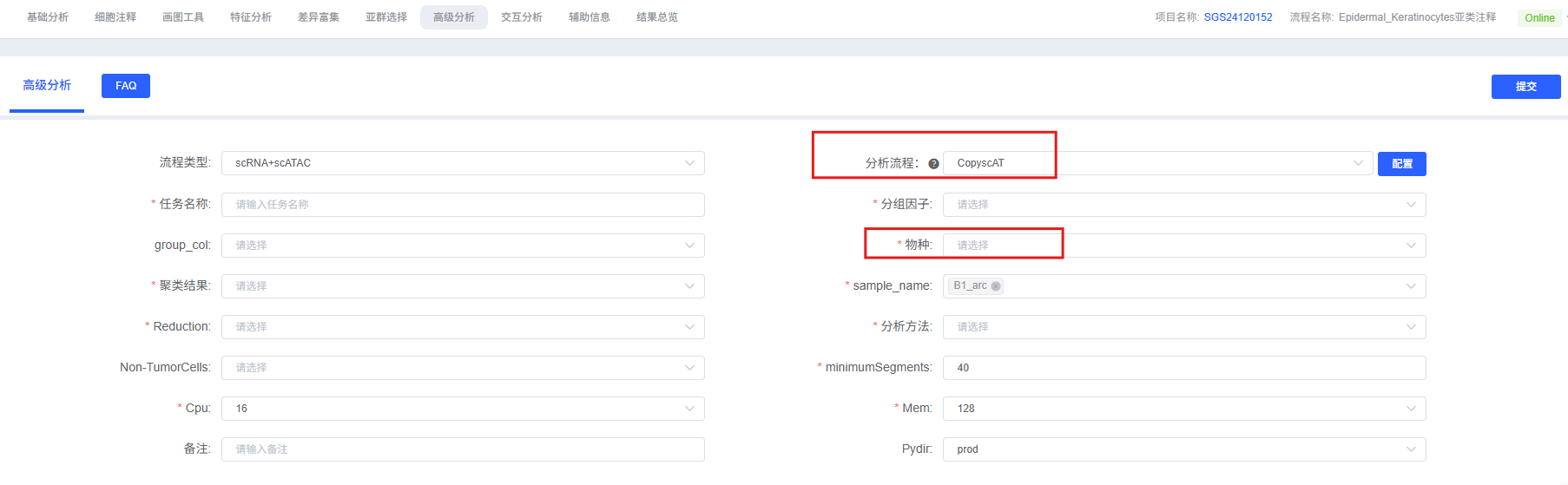
On the cloud platform, the CopyscAT analysis workflow is designed to be intuitive and easy to use. You do not need to write code; you can complete the analysis simply by configuring parameters through the interface.
Preparation Before Analysis
TIP
The success of CopyscAT analysis largely depends on the quality of input data and the sample type. Before starting the analysis, please ensure:
- Data Pre-processing Completed: Your single-cell ATAC data has undergone standard quality control, dimensionality reduction, clustering, and cell type annotation.
- Suitable Sample Type: CopyscAT is mainly applicable to tumor tissue samples; CNV analysis of normal healthy tissues has limited significance.
- Data Quality Requirements: Ensure the fragment file is aligned with cell barcodes and basic QC is completed. Low-quality ATAC data will lead to inaccurate CNV detection.
Parameter Details
The table below details the main parameters and their descriptions for the CopyscAT analysis module on the cloud platform.
| Parameter Name | Description |
|---|---|
| Task Name | The name of this analysis task, must start with an English letter, and can contain English letters, numbers, underscores, and Chinese characters. |
| Group Factor | The result of cell annotation, a column name in metadata, such as CellAnnotation. Used for visualizing cell subgroups. |
| group_col | Sample grouping, a column name in metadata, such as Group. Used for analyzing groups. |
| Species | Species, limited to human or mouse. Can only fill in human or mouse. |
| Clustering Result | Clustering result, a column name in metadata (e.g., wknn_res.0.5_d30_l2_50). Used for cell clustering. |
| sample_name | Sample name, a column name in metadata, such as Sample. |
| reduction | Dimensionality reduction method, including umap/tsne/lsi/atacumap/atactsne/wnnumap/wnntsne, used for FeaturePlot to display CNV distribution of different cell clusters. |
| Analysis Method | Select analysis method, including auto and reference. reference indicates using normal cells as a reference to calculate CNV, while auto does not require reference cells. |
| Non-TumorCells | When Analysis Method selects reference, you need to specify normal cell types used for reference (such as B cells, T cells, etc., immune cells are recommended). |
| minimumSegments | Minimum number of segments: cell filtering threshold, defaults to retaining cells with ≥ 40 segments. |
Important Notes
TIP
- CNV Detection Strategy Selection: CopyscAT provides two CNV detection strategies. Option 1 (using all cells to generate control) is suitable for most cases, especially when the proportion of tumor cells in the sample is high. Option 2 (automatically identifying non-tumor cells as control) is suitable for samples with lower tumor purity (< 90%), capable of setting the baseline more accurately, but note that if the non-tumor cell population is very small, it might be missed.
- Window Size Selection: The choice of window size needs to balance resolution and statistical power. The default 1 Mb is suitable for most cases and can be adjusted according to data characteristics.
- Blacklist Regions: It is strongly recommended to use ENCODE blacklist region files to exclude genomic loci with systematic alignment biases.
- Sample Type: CopyscAT is mainly applicable to tumor tissue samples; CNV analysis of normal healthy tissues has limited significance.
Operation Workflow
- Enter Analysis Module: Navigate to the "Advanced Analysis" module on the cloud platform and select "CopyscAT".
- Create New Task: Name your analysis task and select the sample or project to be analyzed.
- Configure Parameters: According to the guide above, select parameters such as cell type, grouping information, and CNV detection strategy.
- Submit Task: After confirming the parameters are correct, click the "Submit" button and wait for the analysis to complete.
- View Results: After the analysis ends, view the generated analysis report and result files in the task list, including CNV heatmaps, copy number matrices, etc.
Result Interpretation
The CopyscAT analysis report contains rich charts and data files. The following is a detailed interpretation of the core results.
Copy Number Variation Heatmap
The copy number variation heatmap is the core visualization result of CNV analysis, comprehensively displaying the copy number status of all cells across the whole genome:
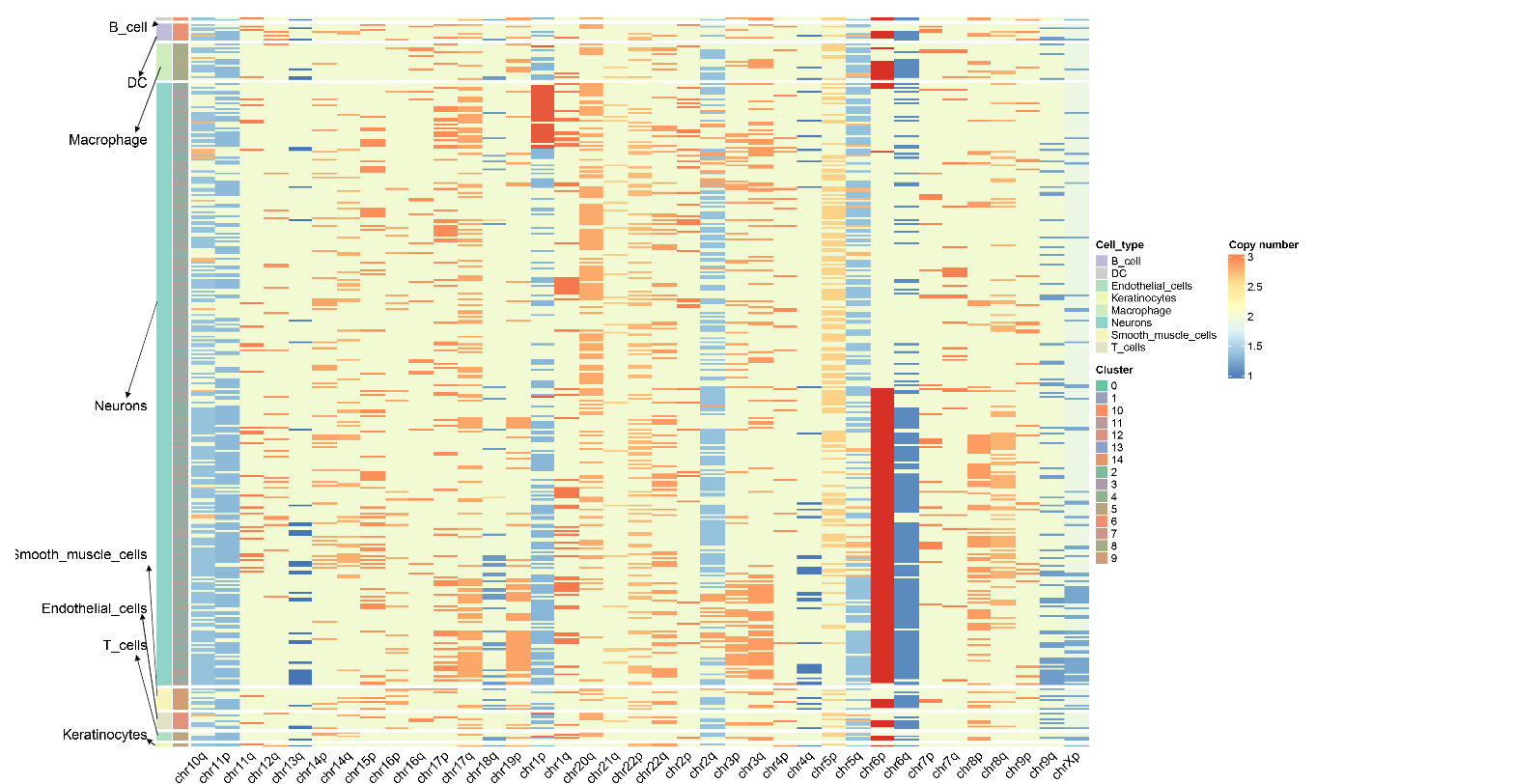
Chart Interpretation
Meaning of Rows and Columns:
- Each row represents a cell.
- Each column represents a chromosome arm, and the color of the heatmap indicates the CNV score.
- The order of columns corresponds to the linear arrangement of the genome from Chromosome 1 to sex chromosomes (if retained).
Color Mapping:
- Copy number value around 2 indicates a normal diploid state.
- Copy number between 1~2 indicates chromatin deletion (loss); the closer to 1, the more severe the deletion.
- Copy number between 2~3 indicates chromatin amplification (gain); the closer to 3, the more severe the amplification.
- Redder colors indicate more severe amplification, and bluer colors indicate more severe deletion.
Cell Annotation:
- The color bands on the left side of the heatmap represent cells arranged by cell type and cell clustering.
- CopyscAT uses consensus clustering methods to finally determine the CNV status and grouping of cells.
- Malignant cells typically exhibit obvious CNV patterns (large-scale amplification or deletion), while non-malignant cells (such as T cells, B cells) maintain a relatively normal diploid state.
Analysis Points
Identify Malignant Cells: By observing CNV patterns, malignant cells can be distinguished from non-malignant cells. Malignant cells typically exhibit large-scale, non-random CNV patterns, while non-malignant cells maintain a relatively normal diploid state.
Resolve Tumor Heterogeneity: Different cell populations may exhibit distinct CNV characteristics. These differences reflect the cellular heterogeneity and subclonal structure within the tumor. CopyscAT is particularly suitable for processing complex samples with high levels of intra-tumor heterogeneity.
Multi-level CNV Detection: CopyscAT can detect local CNV (such as ecDNA), fragment-level CNV, and chromosome-arm-level CNV, providing comprehensive genomic structural variation information.
Result File List
| File/Directory | Content Description |
|---|---|
cnv_plots/chr*_umap.pdf | Visualization of CNV variation for each chromosome arm (* is the chromosome number). |
cnv_heatmap_annotated.pdf | Copy number variation heatmap display, including cell annotation. |
Precautions
1. Importance of Sample Type: CopyscAT is mainly applicable to tumor tissue samples; CNV analysis of normal healthy tissues has limited significance. Before performing CNV analysis, please confirm if the sample type is suitable.
2. Choice of CNV Detection Strategy: CopyscAT provides two CNV detection strategies. Option 1 is suitable for most cases, and Option 2 is suitable for samples with lower tumor purity (< 90%). It is recommended to run with Option 1 first, and try Option 2 if the results are not ideal.
3. Impact of Parameter Selection: The choice of parameters such as window size and filtering thresholds will significantly affect CNV detection results. It is recommended to run with default parameters first, then adjust appropriately based on result quality.
4. Usage of Blacklist Regions: It is strongly recommended to use ENCODE blacklist region files to exclude genomic loci with systematic alignment biases, avoiding interference with copy number inference.
5. Caution in Result Interpretation: CNV detection results need to be interpreted in combination with biological knowledge. Certain technical factors (such as sequencing depth, batch effects) may affect the accuracy of CNV detection.
6. Handling Large Sample Data: For very large datasets, you can subset the data or run analysis on different cell populations separately, then merge the results.
Frequently Asked Questions (FAQ)
Q1: How to choose the appropriate CNV detection strategy?
A: CopyscAT provides two CNV detection strategies:
- Option 1: Uses all cells to generate control. Suitable for most cases, especially when the proportion of tumor cells in the sample is high.
- Option 2: Automatically identifies non-tumor cells as control. Suitable for samples with lower tumor purity (< 90%), capable of setting the baseline more accurately. However, note that if the non-tumor cell population is very small, it might be missed.
Q2: How to interpret values in copy number results?
A: In copy number inference results:
- Close to 2: Indicates normal copy number (diploid state), i.e., the region maintains normal copy number in the corresponding cell.
- Between 1~2: Indicates copy number deletion (loss), the copy number of the region in the corresponding cell is lower than normal level; closer to 1 indicates more severe deletion.
- Between 2~3: Indicates copy number amplification (gain), the copy number of the region in the corresponding cell is higher than normal level; closer to 3 indicates more severe amplification.
- These values reflect the copy number status of each cell in each genomic window, which can be used to identify malignant cells and subclonal structures.
Q3: How to adjust NMF clustering parameters?
A: When using Option 2 (automatically identifying non-tumor cells), if the results are not ideal, you can adjust the following parameters:
- cutHeight: Height for cutting the dendrogram, default is 0.4. If clustering separation is poor, you can lower this value.
- nmfComponents: Number of NMF components, default is 5. Can be adjusted based on sample complexity.
- estimatedCellularity: Estimated proportion of tumor cells, default is 0.8. If the tumor purity of the sample is known, this parameter can be adjusted.
Q4: What are the advantages of CopyscAT compared to other CNV detection tools?
A: The main advantages of CopyscAT include:
- Automatic Identification of Normal Cells: Can automatically identify non-tumor cells in the sample and use them as controls, improving the accuracy of CNV detection.
- Multi-level CNV Detection: Capable of detecting local CNV (such as ecDNA), fragment-level CNV, and chromosome-arm-level CNV.
- Double Minutes Detection: Capable of identifying high copy number amplification regions in the genome (double minutes).
- Cell Cycle Analysis: Provides cell cycle score estimation function, helpful for understanding cell states.
- Suitable for Complex Tumors: Particularly suitable for processing complex samples with high levels of intra-tumor heterogeneity, such as glioblastoma.
Q5: How to handle large sample data?
A: For very large datasets:
- You can subset the data (e.g., using the
subsetSizeparameter). - You can run analysis on different cell populations separately, then merge the results.
- For time-consuming steps like double minutes detection, consider running only on specific cell populations.
Q6: What types of CNV can CopyscAT detect?
A: CopyscAT can detect multiple types of CNV:
- Local CNV: Such as ecDNA (extrachromosomal DNA), inferred through large peaks in the normalized coverage matrix.
- Fragment-level CNV: Infers fragment-level CNV using the normalized matrix.
- Chromosome-arm-level CNV: Infers chromosome-arm-level CNV using the normalized matrix.
- Double Minutes: Identifies high copy number amplification regions in the genome.
References
- Nikolic A, Singhal D, Ellestad K, et al. Copy-scAT: Deconvoluting single-cell chromatin accessibility of genetic subclones in cancer[J]. Science advances, 2021, 7(42):eabg6045. DOI:10.1126/sciadv.abg6045.
