CellPhoneDB
Preface
IMPORTANT
CellPhoneDB is a database and analysis tool for inferring ligand-receptor interactions in human (HUMAN) single-cell transcriptomics data, with special consideration for the structural composition of multi-subunit complexes, suitable for systematic studies of cell-cell communication.
This training document is for bioinformatics analysts and platform users, aiming to introduce the basic principles of CellPhoneDB, parameter configuration and operation workflows on the SeekSoulOnline platform, methods for interpreting output results, as well as a complete case demonstration and notes. Official source code and documentation reference: CellphoneDB GitHub Repository.
CellPhoneDB Introduction and Principles
Core Functions
- Build and use manually curated ligand-receptor databases (HUMAN only)
- Consider subunit structure and complex information of ligands/receptors to accurately model heteromeric complexes
- Provide statistical permutation tests to determine significant ligand-receptor expression between cell type pairs
Analysis Principles (Brief)
- Calculate the average expression and cell expression proportion of ligand or receptor genes in each cell type separately; if a molecule consists of multiple subunits, use the minimum subunit expression to represent the expression of that complex.
- Use random permutation of cell type labels (default 1000 times) to construct null distribution for evaluating the significance of ligand-receptor pairs in two cell types.
- Compare the average expression values of real data with the null distribution. If higher than the specified quantile (e.g., 95%) and meets the expression proportion threshold (default 10%), mark as significant ligand-receptor pairs.
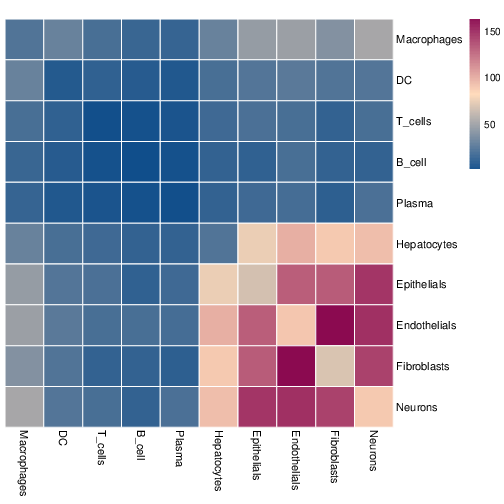
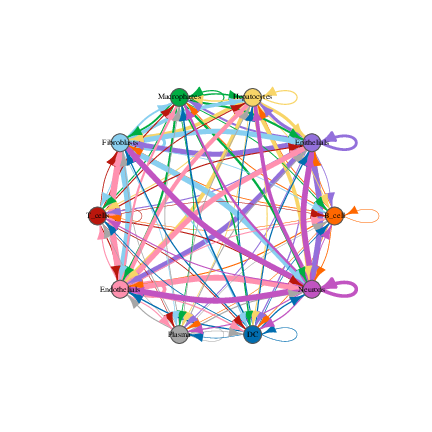
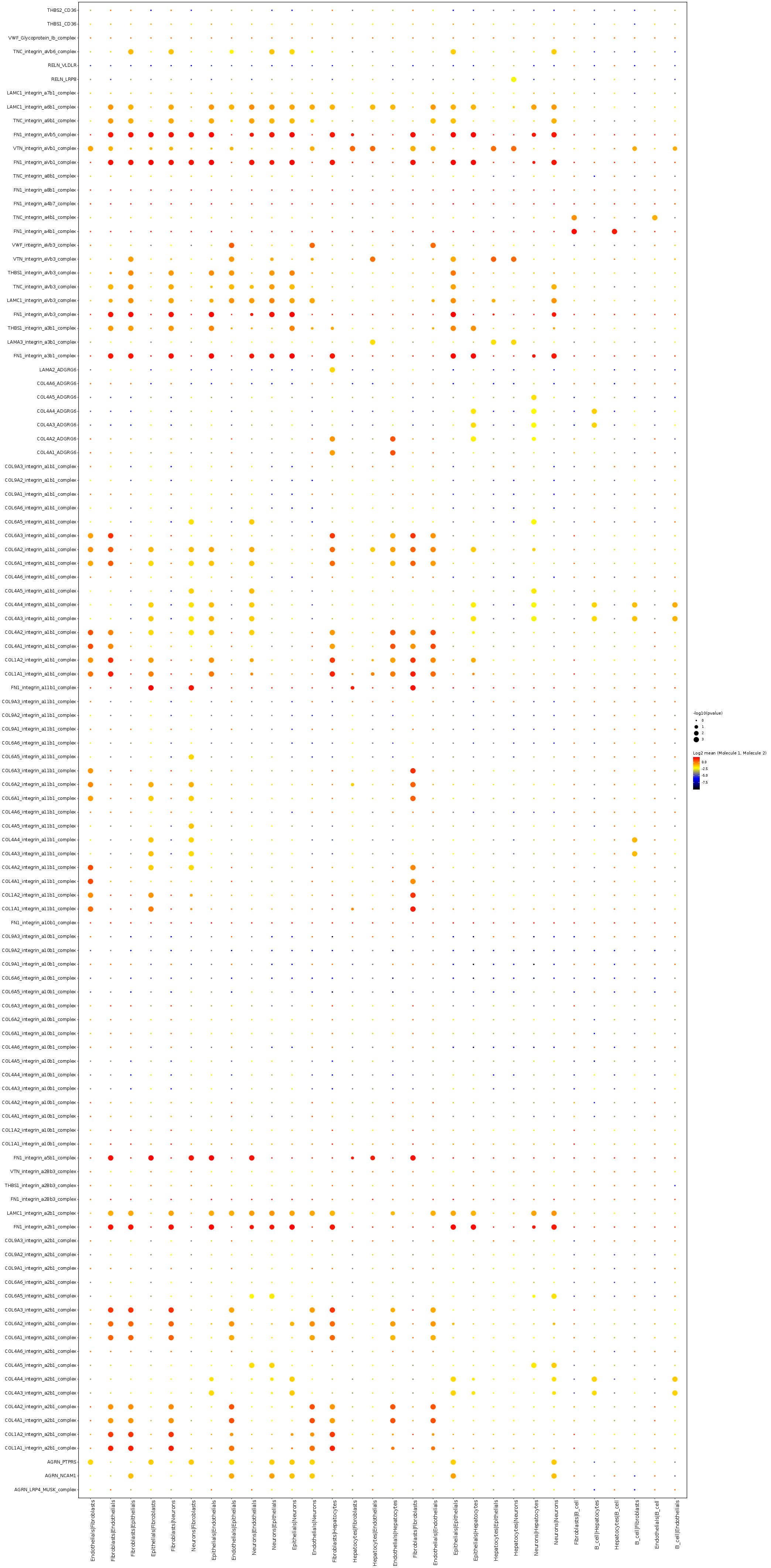
- Summarize significant ligand-receptor pairs into cell-cell interaction matrices, and generate output files such as heatmaps, network plots, ligand-receptor detail tables for downstream interpretation and visualization.
NOTE
The feature of CellPhoneDB is "subunit/complex sensitivity", which can significantly reduce misjudgment when handling heteromeric receptors or polypeptide ligands.
SeekSoulOnline Platform Parameters and Operation Guide
The following are recommended parameters and their meanings when running CellPhoneDB analysis on the SeekSoulOnline platform (platform UI field examples).
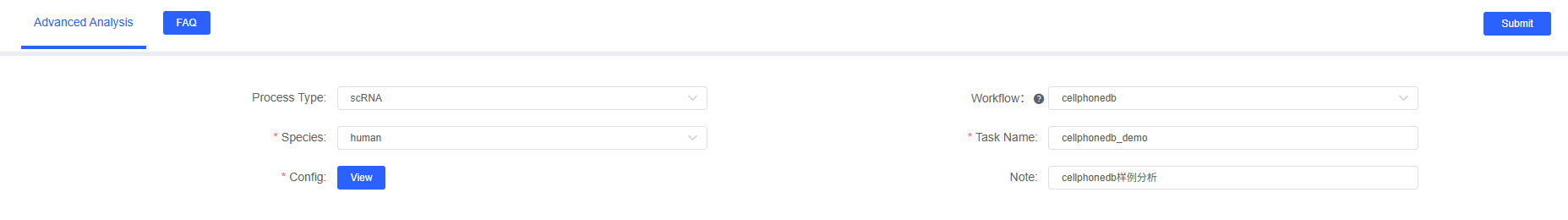
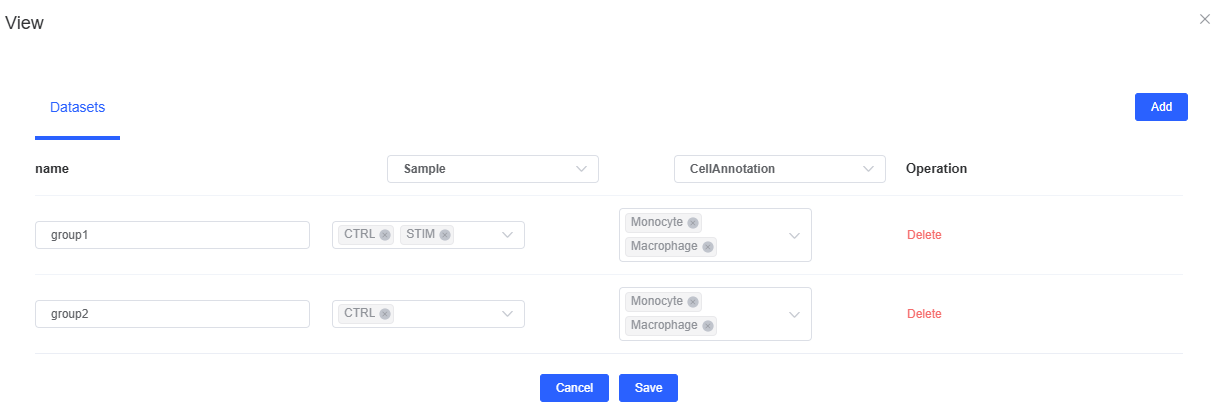
Basic Parameters
- Task Name: Starts with English letters, allows letters, numbers, underscores, and Chinese, for example
cellphonedb_analysis_2025 - Species: Select
humanormouse. - Config: After configuring the first row, click [Add], customize group names in the first column, select samples (multiple selection allowed) in the second column, and select cell types (multiple selection allowed) in the third column.
Platform Operation Workflow (Brief)
- Configure parameters and submit task
- Wait for analysis to complete, download results and perform visualization and report writing on the platform or locally
Analysis Workflow and Example Output
Core Output Files
all_count_network.txt: Cell-cell interaction ligand-receptor pairsdeconvoluted.txt: Decomposed expression information of ligand-receptor molecules in cell types, with the last few columns being the average expression of genes in each cell typemeans.txt: Average values of ligand-receptor pair interactionspvalues.txt: P-value table obtained from permutation tests- Visualization images: Heatmaps, network plots, Circos/Chord plots, etc. (see examples below)
Case Demonstration
Cell-cell interaction strength heatmap:
Cell-cell interaction network plot (example):
Top ligand and receptor interaction subsets (example bubble plot/heatmap):
Notes and Best Practices
- Species Limitations: CellPhoneDB database is mainly HUMAN; mouse data needs gene name mapping before analysis.
- Cell Numbers and Statistical Power: Each cell type is recommended to have at least 50 cells (rare cells can be relaxed, but need careful interpretation).
- Threshold Selection: Too low expression thresholds may increase false positives, too high may miss real signals.
- Complex Handling: Pay attention to checking the expression of all subunits of complexes to avoid misjudging entire complex activity due to single subunit bias.
- Result Validation: It is recommended to combine differential expression, enrichment analysis, and experimental validation (such as in vitro functional experiments or tissue section in situ hybridization).
Frequently Asked Questions (FAQ)
Q1: Does CellPhoneDB support non-human species?
A1: The official database is HUMAN, other species need to map gene names first before running.
Q2: Can the number of permutations be reduced to speed up?
A2: Yes, but it will reduce the stability of p-value estimation. For exploratory analysis, the number of permutations can be appropriately reduced.
Q3: How to handle expression differences of individual molecules in complexes?
A3: CellPhoneDB uses the minimum subunit expression to represent complex expression. Manual checking of each subunit expression is needed to confirm conclusions.
References
- CellPhoneDB official repository:
https://github.com/ventolab/CellphoneDB