FFPE Single-Cell Data Processing Case and Optimization Recommendations
Case Background
- Sample Type: Gastric cancer FFPE (formalin-fixed paraffin-embedded) tissue
- Problem Description: The original single-cell expression matrix, when directly integrated and clustered, showed unsatisfactory clustering results, unclear cluster separation, difficulty in cell type annotation, and significant background RNA interference.
Data Processing and Optimization Workflow
1. Preprocessing and Quality Control
- Perform initial quality control on the expression matrix of each sample.
- Use SoupX to remove background RNA contamination.
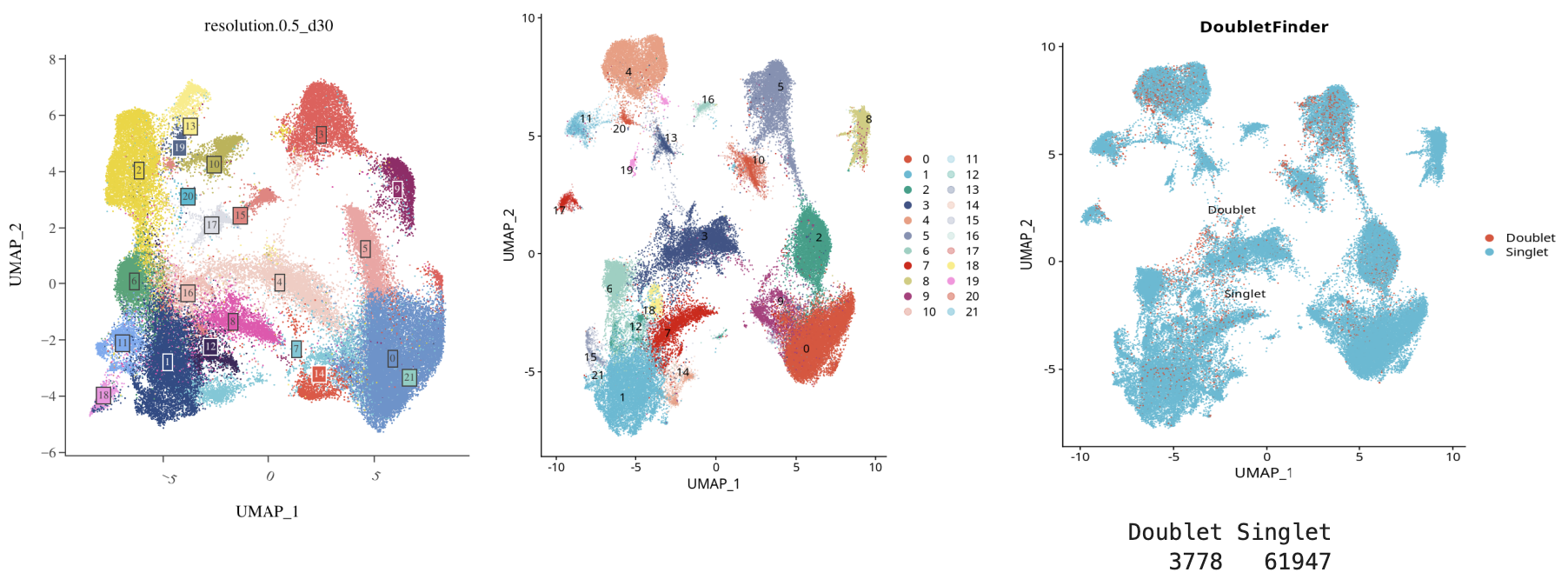
- Use DoubletFinder to filter doublets, with the following thresholds:
- UMI count: 500-75,000
- Gene count: 200-7,500
- Mitochondrial gene ratio: <10%
TIP
Due to severe RNA degradation and background contamination in FFPE samples, it is strongly recommended to perform background removal and doublet filtering.
2. Clustering and Annotation Effect Comparison
- Before processing: Clusters are not clearly separated, and cell types are difficult to annotate accurately.
- After processing:
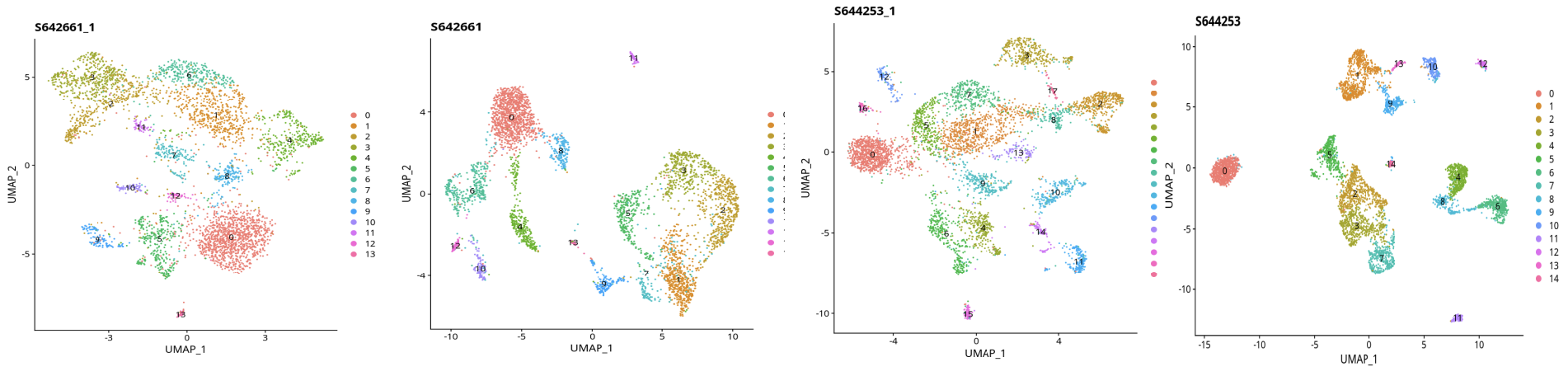
- After background removal with SoupX, cluster boundaries become clear and clustering improves significantly.
- DoubletFinder further removes doublets, improving clustering purity.
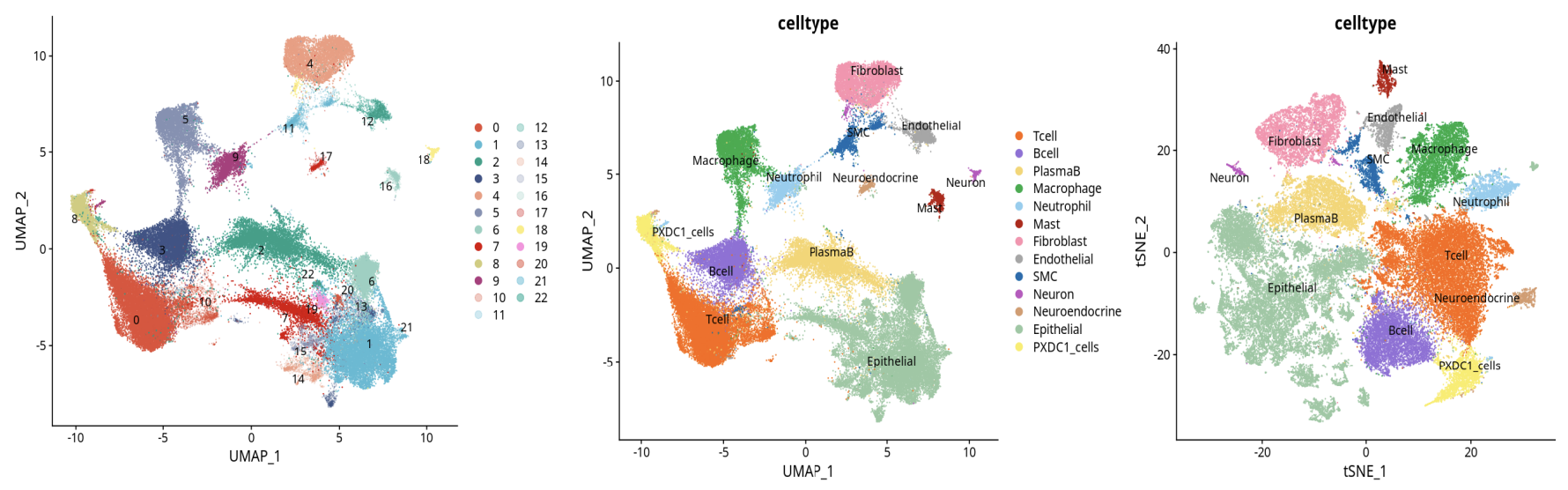
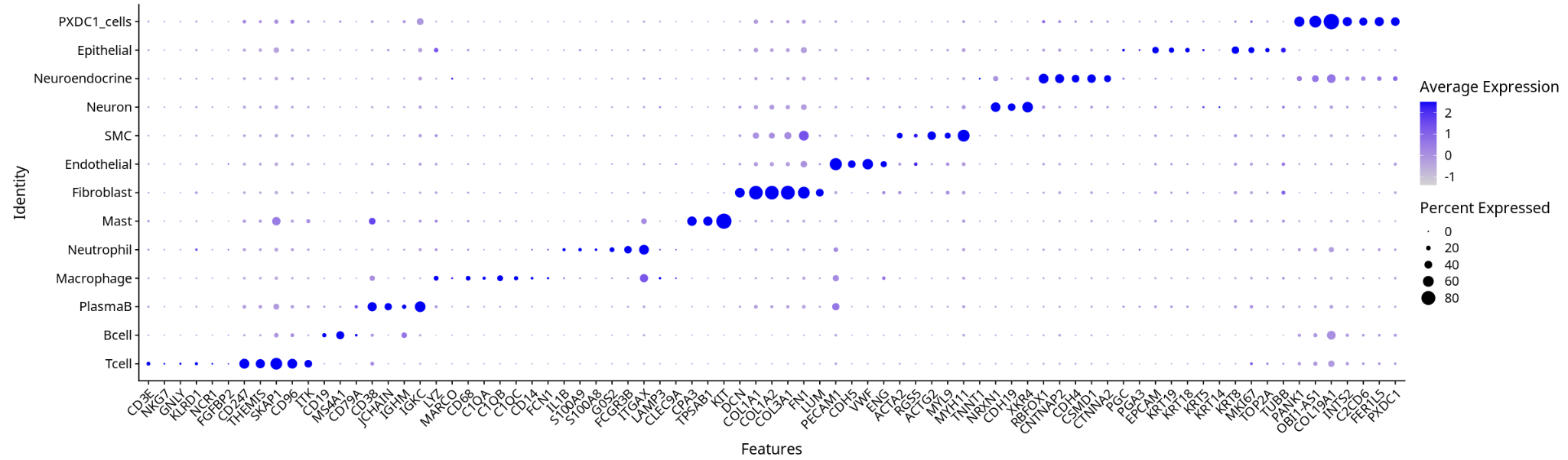
- Cell type annotation and marker gene expression distribution better match biological expectations.
Caption: Left is the original clustering, right is clustering after SoupX processing
Caption: Left is the original clustering, middle is after SoupX processing, right is after DoubletFinder doublet removal (doublet rate ~6%)
3. Cell Type Annotation and Marker Gene Expression
4. Experience Summary and Optimization Recommendations
IMPORTANT
Common issues with FFPE samples include severe RNA degradation, low total RNA, short fragments, poor nuclear integrity, low nuclear rate (20-30%), and high background RNA, leading to diffuse clustering.
- Data processing recommendations:
- Background RNA removal is essential (e.g., SoupX)
- Doublet filtering (e.g., DoubletFinder)
- Strict quality control (UMI, gene count, mitochondrial ratio)
- Feature selection of highly variable genes, optimize clustering parameters (e.g., number of PCs, resolution)
- Batch integration is recommended to use methods like Harmony for parameter optimization
- After clustering, manually adjust subpopulations based on marker genes and remove mixed populations
NOTE
Improvement in clustering and annotation depends on multi-step optimization. It is recommended to flexibly adjust parameters according to sample characteristics.
Conclusion
This case demonstrates an optimized processing workflow for FFPE and other RNA expression library samples with unsatisfactory clustering. Through background removal, doublet filtering, and parameter optimization, clustering and annotation effects can be significantly improved. This workflow can serve as a reference and recommendation for similar issues, helping users improve the quality of single-cell analysis for FFPE and complex samples.