Surface Protein Antibody and CRISPR QC Analysis
Time: 4 min
Words: 655 words
Updated: 2025-08-08
Reads: 0 times
This workflow applies to QC and preliminary analysis of DD Surface Protein (Antibody) and CRISPR data.
WARNING
SeekSoul Tools does not natively support these two library types. Convert the data to 10x format first, then use Cell Ranger to complete the analysis. Please refer to 10x Genomics’ official tutorials for detailed instructions and interpretation of results.
1. Surface Protein (Antibody)
1. Joint analysis with transcriptome and VDJ libraries
- fastp data QC (optional)
- Data conversion:
- Convert 5' data first to generate
map.txt(mapping between SeekOne barcodes and 10x barcodes). - Then convert Antibody and VDJ data, following the barcode mapping in the successfully converted 5' data
map.txt. - See
Barcode-Converter.zipfor the conversion tool; the archive contains detailed usage instructions.
- Convert 5' data first to generate
- Cell Ranger multi analysis
- Command:
bash
cellranger multi \
--id= sample \
--csv=multi-config-template-7.1-antigen.csv \
--localcores 8 \
--localmem 64- Notes:
multi-config-template-7.1-antigen.csv: configuration file.- feature file: the captured surface protein feature file, provided by the customer. Format as follows:
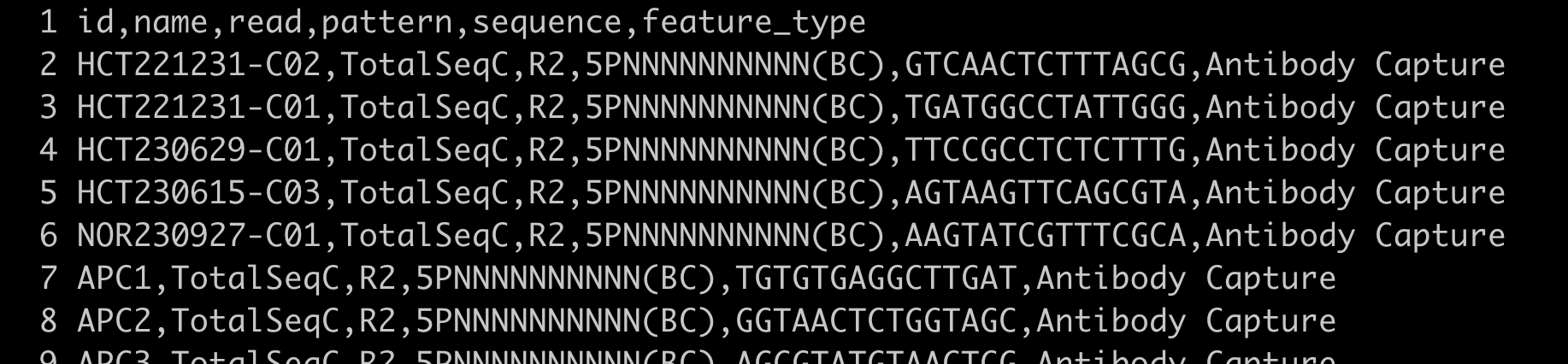
2. Standalone count analysis for the Antibody library
- fastp data QC (optional)
- Convert Antibody data directly to 10x format.
- Cell Ranger count analysis
- Command:
bash
cellranger count \
--id= Sample \
--libraries=library.csv \
--transcriptome=/PROJ2/DATA/database/refdata-gex-GRCh38-2020-A \
--feature-ref=change_Reference.csv \
--localcores 8 \
--localmem 64- Notes:
library.csv: configuration file.- feature file: the captured surface protein feature file, provided by the customer; the format is the same as in the joint analysis.
2. CRISPR
1. Joint analysis with transcriptome library
- fastp data QC (optional)
- Data conversion:
- Convert 5' data first to generate
map.txt(mapping between SeekOne barcodes and 10x barcodes). - Then convert CRISPR data, following the barcode mapping in the successfully converted 5' data
map.txt. - See
Barcode-Converter.zipfor the conversion tool; the archive contains detailed usage instructions.
- Convert 5' data first to generate
- Cell Ranger analysis parameters:
bash
cellranger count \
--id=G1_1 \
--libraries=G1_1.library.csv \
--transcriptome=feature.ref.final_change_final_rev_new_pattern.csv \
--localcores=8 \
--localmem=64Notes:
G1_1.library.csv: configuration file; can include transcriptome, VDJ, and CRISPR data simultaneously.feature-ref: captured gene feature sequences; prepare according to 10x Genomics’ official guidance (format shown below).pattern: the capture feature recognition sequence located upstream of the feature sequence.feature_type: reverse complementary sequence of the gene capture sequence provided by the customer.

text
Example of library.csv:
fastqs,sample,library_type
data/5data/,G1_1_5,Gene Expression
data/crispr_data/,G1_1_crispr,CRISPR Guide Capture2. Joint analysis with transcriptome and VDJ libraries
- fastp data QC (optional)
- Data conversion:
- Convert 5' data first to generate
map.txt(mapping between SeekOne barcodes and 10x barcodes). - Then convert CRISPR and VDJ data, following the barcode mapping in the successfully converted 5' data
map.txt. - See
Barcode-Converter.zipfor the conversion tool; the archive contains detailed usage instructions.
- Convert 5' data first to generate
- Cell Ranger multi analysis:
bash
cellranger multi \
--id=G1_5 \
--csv=multi-config.csv \
--localcores 8text
Format of multi-config.csv configuration file:
[gene-expression]
reference,/database/refdata-gex-GRCh38-2020-A
include-introns,true
[feature]
reference,crispr_Reference.csv
[vdj]
reference,database/refdata-cellranger-vdj-GRCh38-alts-ensembl-5.0.0
[libraries]
fastq_id,fastqs,lanes,feature_types,subsample_rate
G1_5_5,CRISPR/demo/data/,,Gene Expression,1,
G1_5_crispr,CRISPR/demo/data/,,CRISPR Guide Capture,1,
G1_5_BCR,CRISPR/demo/data/,,VDJ-B,1,text
Format of crispr_Reference.csv (CRISPR feature configuration file):
TIP
Before conversion and analysis, verify that the map.txt mapping matches the feature reference files. After conversion, consider running a small-scale pilot to validate the setup before processing the full dataset.
