GFP Transgene Not Detected
Table of Contents
Problem Description
The customer found that although the GFP transgene was included in the reference genome, the expression of the GFP gene could not be detected in the cloud platform analysis results.
Troubleshooting Process
- Initial Confirmation:
- The expression matrix contained the GFP gene, but the RDS file did not, resulting in no corresponding gene in the cloud platform analysis.
- Sample Verification:
- For example, in one sample, the BAM file could be aligned to the GFP gene, and all alignments were 150 matches.
- However, the
XStag was alwaysXS:Z:Unassigned_NoFeatures. - This indicated that the GFP gene could be aligned, but was not recognized during quantification by featureCounts.
zcat features.tsv.gz | grep -n GFP
zcat matrix.mtx.gz | awk -v gene_row=index '$1 == gene_row { count++ } END { print count }'
samtools view sample_SortedByCoordinate_withTag.bam | grep "GFP"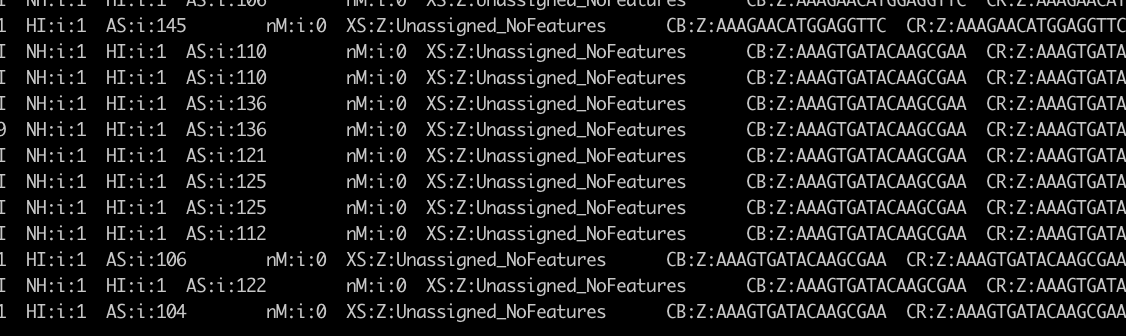
- Multiple Sample Review:
- Other samples showed the same issue: the GFP gene was not quantified.
Cause Analysis
IMPORTANT
The main reason why the GFP gene was not recognized by featureCounts is that the GTF file format was non-standard: the annotation for the exogenous GFP gene lacked gene and transcript lines, containing only exon information. This prevented featureCounts from quantifying it correctly.
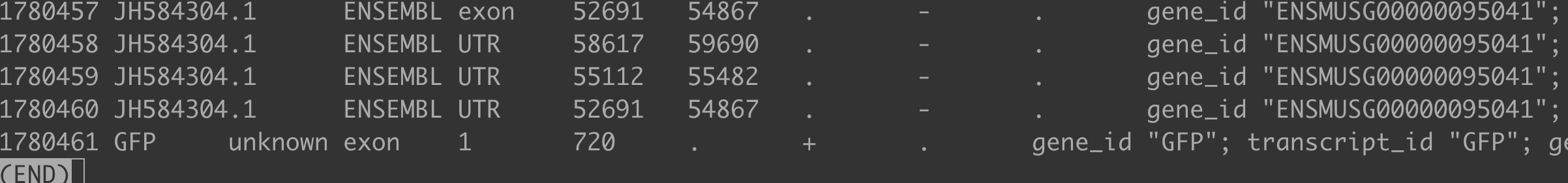
Solution
TIP
You need to reconstruct the GTF annotation file for the GFP gene, ensuring that it contains all three annotation lines: gene, transcript, and exon.
Standard GFP Gene GTF Format Example:
# Add gene line
echo -e 'GFP\tunknown\tgene\t1\t720\t.\t+\t.\tgene_id "GFP"; gene_name "GFP"; gene_biotype "protein_coding";' > GFP.gtf
# Add transcript line (append with >>)
echo -e 'GFP\tunknown\ttranscript\t1\t720\t.\t+\t.\tgene_id "GFP"; transcript_id "GFP"; gene_name "GFP"; gene_biotype "protein_coding";' >> GFP.gtf
# Add exon line (append with >>)
echo -e 'GFP\tunknown\texon\t1\t720\t.\t+\t.\tgene_id "GFP"; transcript_id "GFP"; gene_name "GFP"; gene_biotype "protein_coding"; exon_number 1; exon_id "GFP"' >> GFP.gtf
NOTE
After reconstructing the reference genome with the corrected GTF file and re-running quantification, the GFP gene can be properly detected and quantified.
