Overview
SeekOne™ DD Single Cell Full-length RNA Sequence Transcriptome-seq kit
—— The Time Has Come for A New Generation of Single Cell Transcriptome Profiling
SeekOne™ Digital Droplet (SeekOne™ DD) High throughput Single Cell Full-length RNA Sequence Transcriptome-seq (scFAST-seq) Kit, self- developed by Beijing SeekGene BioSciences Co., Ltd., is a powerful commercial tool for high-throughput whole transcriptome profiling. Compared to conventional 3' scRNA-seq, scFAST- seq has distinct advantages in detecting non- polyadenylated transcripts, transcript coverage length, and identification of more splice junctions. With target region enrichment, scFAST-seq can simultaneously detect somatic mutations and cell states in individual tumour cells, providing valuable information for precision medicine.
SeekOne™ DD Single Cell Full-length RNA Sequence Transcriptome-seq (scFAST-seq) Kit includes SeekOne™ DD Chip S3 (Chip S3), Gasket, Carrier Oil, SeekOne™ DD scFAST-seq Barcoded Beads, amplification reagents, library construction reagents.
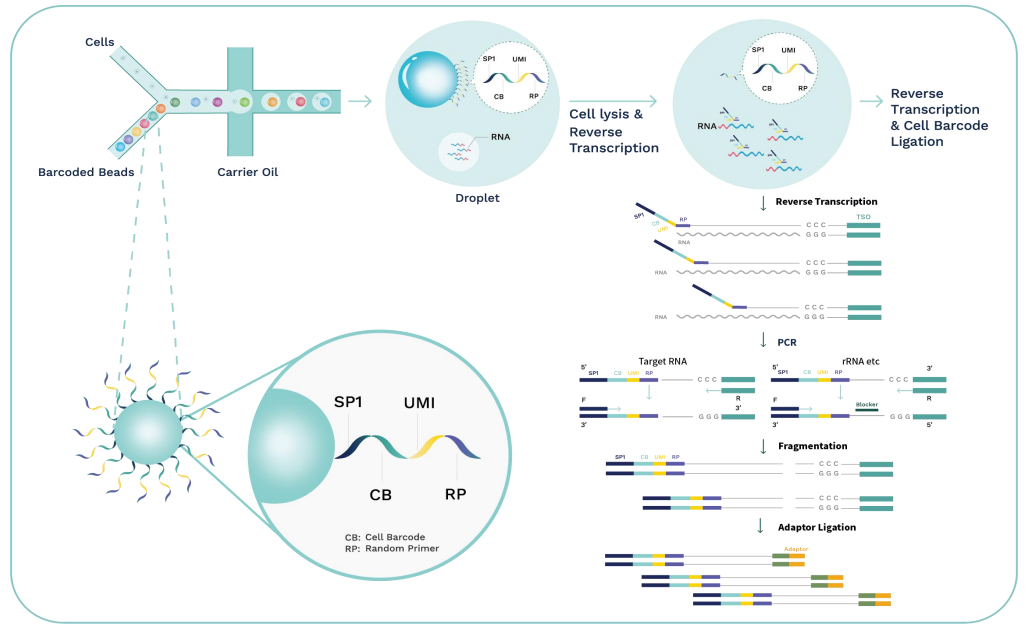
Core technology and workflow
Using microfluidic digital droplets and Barcoded Beads coupled with random primers, single-cell partition and random capture of the whole transcriptome can be achieved in a step of generating water-in-oil droplets. This convenient process requires only a small amount of sample. The inclusion of an rRNA blocker enables efficient depletion of rRNA, delivering higher proportion of effective data.
- 1-8 samples can be flexibly run at a time
- 500-12,000 cells can be captured with a single channel
- 150,000 emulsion droplets can be generated in 3 minutes
- Recovery of ~1000 cells with doublet rates of ~0.3% per 1,000 cells
- Even coverage of the full-length of the gene from 5' to 3'end
- Flexibility to incorporate a panel for detecting target genes with high
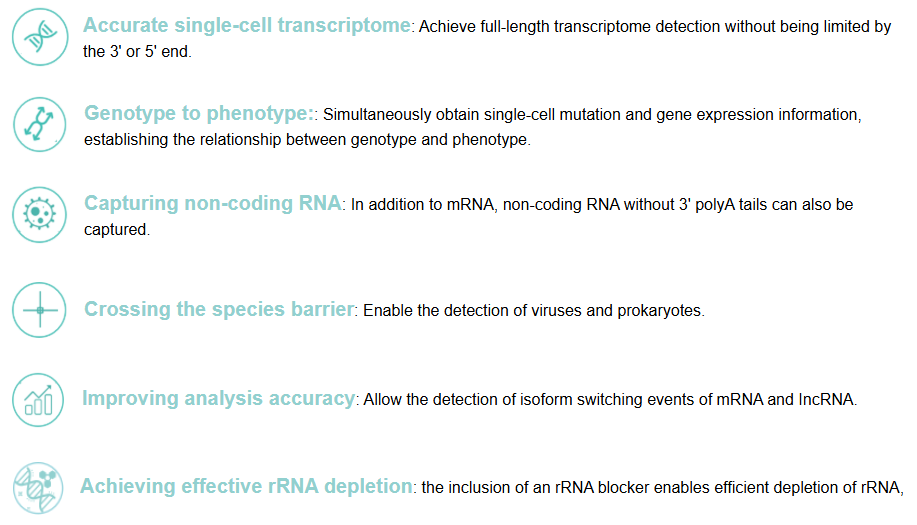
Product Features
Applications
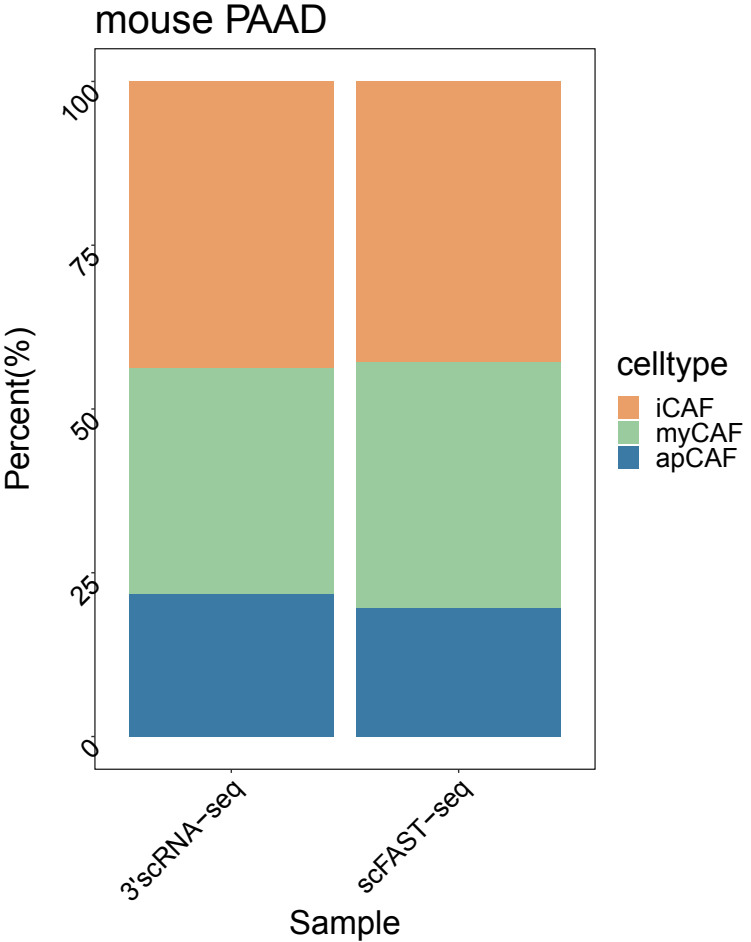
Compared to conventional 3' scRNA-seq, scFAST- seq has distinct advantages in detecting non- polyadenylated transcripts, transcript coverage length, and identification of more splice junctions. With target region enrichment, scFAST-seq can simultaneously detect somatic mutations and cell states in individual tumour cells, providing valuable information for precision medicine.
Detection of single cell mutation
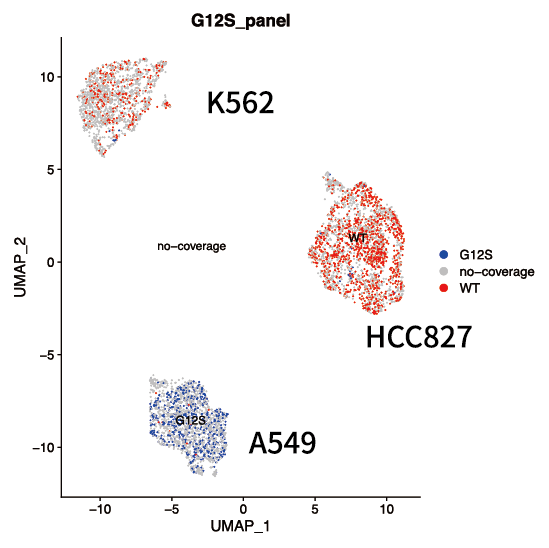
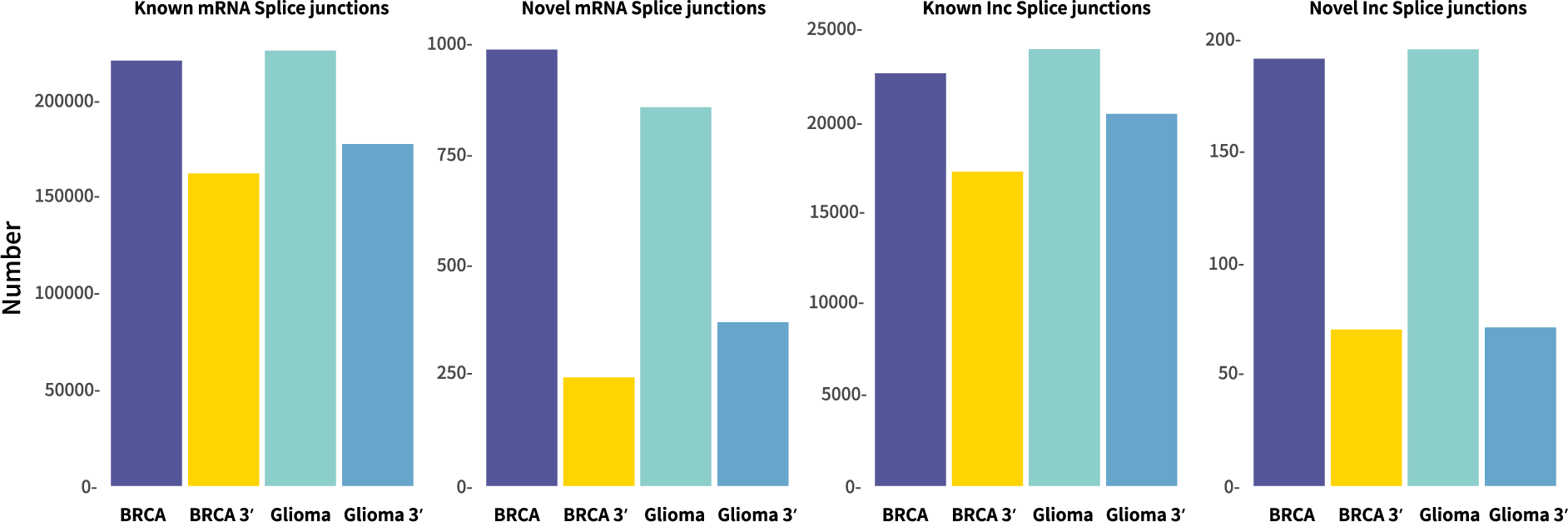
Detection of more splice junctions
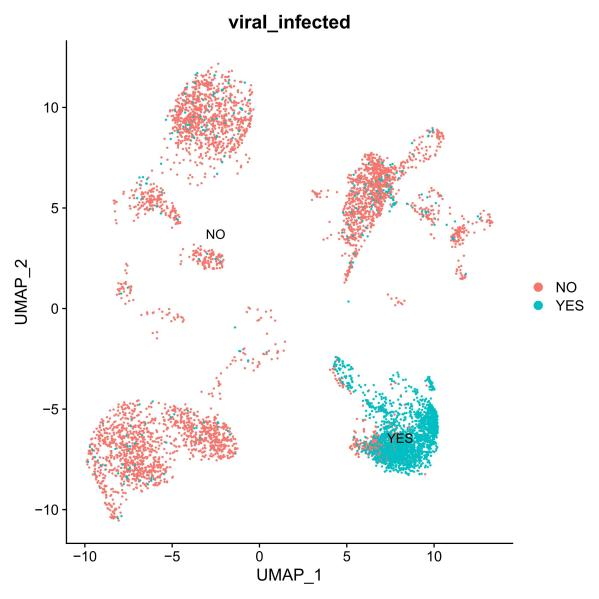
Identification of non-polyA tailed viral infected cell clusters
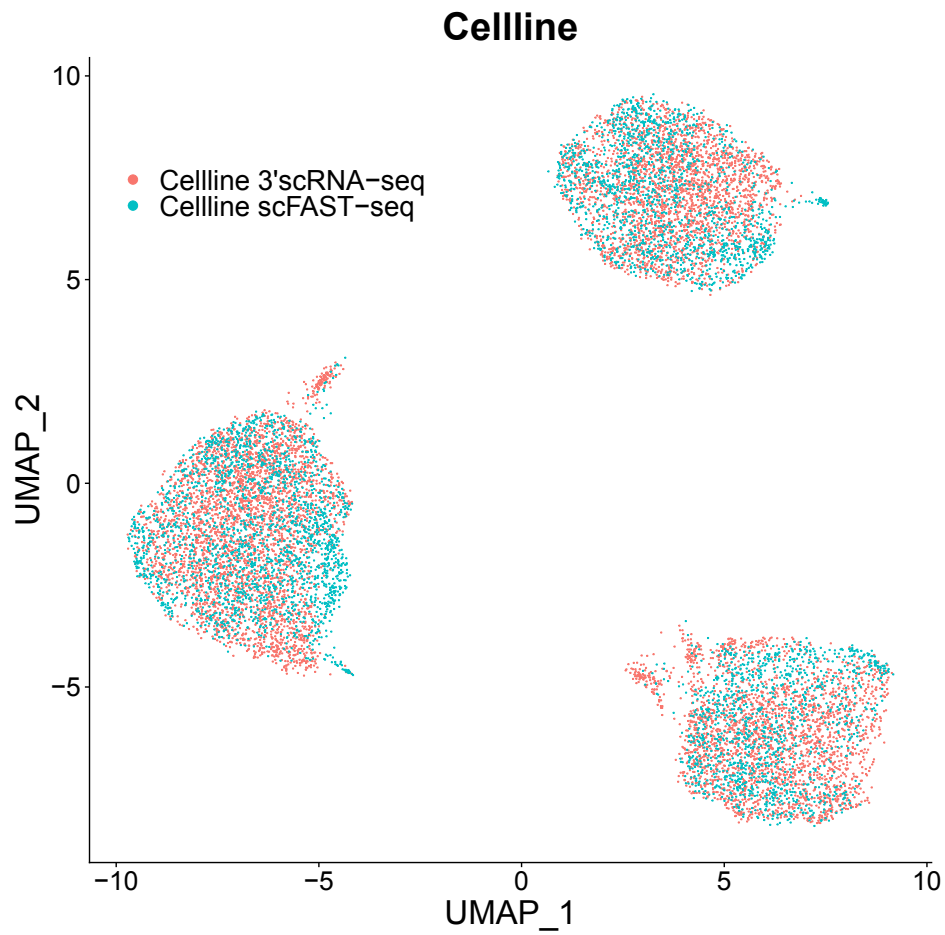
Clearer and more accurate cell trajectory inference compared to the 3' platform

scFAST-seq data can be integrated with 3' data