SeekSoul Tools v1.0.0
SeekSoulTools is a software developed by SEEKGENE for processing single-cell transcriptome data. It is used for identifying cell barcode, genome alignment and gene quantification to obtain a feature-barcode matrix that can be used for downstream analsis, followed by cell clustering and differential analysis. This module not only support data from SeekOne transcriptome kits, it also supports a variety of customized structure designs.
Download
SeekSoul Tools v1.0.0 Download-SeekSoulTools - md5: d2724dc8213ec8e9d3b3872caa71c585
wget
mkdir seeksoultools.1.0.0
cd seeksoultools.1.0.0
wget -c -O seeksoultools.1.0.0.tar.gz "https://seekgene-public.oss-cn-beijing.aliyuncs.com/software/seeksoultools/seeksoultools.1.0.0.tar.gz"curl
mkdir seeksoultools.1.0.0
cd seeksoultools.1.0.0
curl -C - -o seeksoultools.1.0.0.tar.gz "https://seekgene-public.oss-cn-beijing.aliyuncs.com/software/seeksoultools/seeksoultools.1.0.0.tar.gz"Installation Guide
Installation:
IMPORTANT
Please ensure you follow these steps correctly to install the software, otherwise functionality may be affected.
# decompress
tar zxf seeksoultools.1.0.0.tar.gz
# install
source ./bin/activate
./bin/conda-unpack
# export path in bashrc
export PATH=`pwd`/bin:$PATH
echo "export PATH=$(pwd)/bin:\$PATH" >> ~/.bashrcConfirm installation:
seeksoultools --versionTutorials
Data preparation
NOTE
Before starting the analysis, please make sure you have prepared the following required files:
- Sequencing data (FASTQ format)
- Reference genome for the corresponding species
- Gene annotation file (GTF format)
Download sample datasets
sample datasets - md5: 6601c8f3c4c827d2a30fcb5c6d0dee7c(Species: Homo sapiens.)
wget
wget -c -O demo3k.tar "https://seekgene-public.oss-cn-beijing.aliyuncs.com/software/data/demodata/demo3k.tar"
# decompress
tar xf demo3k.tarcurl
curl -C - -o demo3k.tar "https://seekgene-public.oss-cn-beijing.aliyuncs.com/software/data/demodata/demo3k.tar"
# decompress
tar xf demo3k.tarDownload and build reference genome
Download-human-reference-GRCh38 - md5: 5473213ae62ebf35326a85c8fba6cc42
wget
mkdir -p /demo/refdata/
cd /demo/refdata/
wget -c -O GRCh38.tar.gz "https://seekgene-public.oss-cn-beijing.aliyuncs.com/software/data/reference/GRCh38.tar.gz"
# decompress
tar -zxvf GRCh38.tar.gzcurl
mkdir -p /demo/refdata/
cd /demo/refdata/
curl -C - -o GRCh38.tar.gz "https://seekgene-public.oss-cn-beijing.aliyuncs.com/software/data/reference/GRCh38.tar.gz"
# decompress
tar -zxvf GRCh38.tar.gzThe assembly of the reference genome refers to How to build reference genome?
Run Seeksoultools
Run tests
TIP
SeekSoulTools provides various running modes. The examples below cover the most common usage scenarios. You can choose the appropriate parameter combinations according to your needs.
Example 1: Basic usage
mkdir -p /demo/myproject/
cd /demo/myproject/
seeksoultools rna run \
--fq1 /demo/data/demo3k_R1_001.fastq.gz \
--fq2 /demo/data/demo3k_R2_001.fastq.gz \
--samplename demo3k \
--outdir /demo/myproject/ \
--genomeDir /demo/refdata/GRCh38-3.0.0/star \
--gtf /path/demo/refdata/GRCh38-3.0.0/genes/genes.gtf \
--chemistry MM \
--core 4Example 2: Specify a different version of STAR for analysis. Make sure that the STAR version is compatible with the –genomeDir.
IMPORTANT
When using a custom STAR version, you must ensure it is compatible with the version used to build the genome index, otherwise the analysis will fail.
mkdir /demo/myproject/
cd /demo/myproject/
seeksoultools rna run \
--fq1 /demo/data/demo3k_R1_001.fastq.gz \
--fq2 /demo/data/demo3k_R2_001.fastq.gz \
--samplename demo3k \
--outdir /demo/myproject/ \
--genomeDir /demo/refdata/GRCh38/star \
--gtf /path/demo/refdata/GRCh38/genes/genes.gtf \
--chemistry MM \
--core 4 \
--star_path /path/to/cellranger-5.0.0/lib/bin/STARExample 3: A sample has multiple sets of fastq files
mkdir /demo/myproject/
cd /demo/myproject/
seeksoultools rna run \
--fq1 /demo/data/demo_S1_L001_R1_001.fastq.gz \
--fq1 /demo/data/demo_S1_L002_R1_001.fastq.gz \
--fq2 /demo/data/demo_S1_L001_R2_001.fastq.gz \
--fq2 /demo/data/demo_S1_L002_R2_001.fastq.gz \
--samplename demo \
--outdir /demo/myproject/ \
--genomeDir /demo/refdata/GRCh38/star \
--gtf /demo/refdata/GRCh38/genes/genes.gtf \
--chemistry MM \
--core 4Example 4: Customize the structure of R1
seeksoultools rna run \
--fq1 /demo/data/demo3k_R1_001.fastq.gz \
--fq2 /demo/data/demo3k_R2_001.fastq.gz \
--samplename demo \
--outdir /demo/myproject/ \
--genomeDir /demo/refdata/GRCh38/star \
--gtf /demo/refdata/GRCh38/genes/genes.gtf \
--barcode /demo/utils/CLS1.txt \
--barcode /demo/utils/CLS2.txt \
--barcode /demo/utils/CLS3.txt \
--linker /demo/utils/Linker1.txt \
--linker /demo/utils/Linker2.txt \
--structure B9L12B9L13B9U8 \
--core 4NOTE
- The structure of read1 is represented by
B9L12B9L13B9U8, which means it consists of three sections of cell barcode, each with 9 bases, and a UMI section with 8 bases. The linker section between the cell barcode and UMI consists of two parts, with the first part being 12 bases and the second part being 13 bases - Use
--barcodeto specify the three sections of barcodes sequentially, and use--linkerto specify the two sections of linkers sequentially.
Parameter descriptions
IMPORTANT
The following parameters have a significant impact on analysis results. Please select carefully based on your experimental design and data characteristics:
- --chemistry: Must exactly match the type of kit used
- --include-introns: Affects gene expression quantification strategy
- --expectNum: Affects the estimated number of cells
| Parameters | Descriptions |
|---|---|
| --fq1 | Paths to R1 fastq files. |
| --fq2 | Paths to R2 fastq files. |
| --samplename | Sample name. A directory will be created named after the sample name in the outdir directory. Only digits, letters, and underscores are supported. |
| --outdir | Output directory. Default: ./ |
| --genomeDir | The path of the reference genome generated by STAR. The version needs to be consistent with the STAR used by seeksoultools. |
| --gtf | Path to the GTF file for the corresponding species. |
| --core | Number of threads used for the analysis. |
| --chemistry | Reagent type, with each type corresponding to a combination of--shift,--pattern, --structure, --barcode, and --sc5p,Available options: DDV1, DDV2, DD5V1, MM, MM-D, DD-Q. DDV1 corresponds to the 3' transcriptome-seq kit V1 reagent for the DD platform. DDV2 corresponds to the 3' transcriptome-seq kit V2 reagent for the DD platform. DD5V1 corresponds to the 5' transcriptome-seq kit V1 reagent for the DD platform. MM corresponds to the 3' transcriptome-seq kit reagent for the MM platform. MM-D corresponds to the large-well transcriptome-seq kit for the MM platform. DD-Q corresponds to the full-length rna sequence transcriptome-seq kit for the DD platform. |
| --skip_misB | If enabled, no base mismatch is allowed for barcode. Default is 1. |
| --skip_misL | If enabled, no base mismatch is allowed for linker. Default is 1. |
| --skip_multi | If enabled, discard reads that can be corrected to multiple white-listed barcodes. Barcodes are corrected to the barcode with the highest frequency by default. |
| --expectNum | Estimated number of captured cells. |
| --forceCell | When number of cells obtained from analysis is abnormal, add this parameter with expected value N. Seeksoultools will select the top N cells based on UMI from high to low. |
| --include-introns | When disabled, only exon reads are used for quantification. When enabled, intron reads are also used for quantification. |
| --star_path | Path to another version of STAR for alignment. The version must be compatible with the--genomeDirversion. The default--star_pathis the STAR in the environment. |
Output descriptions
./
├── demo_report.html 1
├── demo_summary.csv 2
├── demo_summary.json
├── step1
│ └──demo_2.fq.gz
├── step2
│ ├── featureCounts
│ │ └── demo_SortedByName.bam
│ └── STAR
│ ├── demo_Log.final.out
│ └── demo_SortedByCoordinate.bam
├── step3
│ ├── filtered_feature_bc_matrix 3
│ └── raw_feature_bc_matrix
└──step4
├── FeatureScatter.png
├── FindAllMarkers.xls
├── mito_quantile.xls
├── nCount_quantile.xls
├── nFeature_quantile.xls
├── resolution.xls
├── top10_heatmap.png
├── tsne.png
├── tsne_umi.png
├── tsne_umi.xls
├── umap.png
└── VlnPlot.png- Final report in html
- Quality control information in csv
- Filtered feature-barcode matrix
Algorithms Overview
step1: barcode/umi extraction
CAUTION
Accurate extraction of barcodes and UMIs is critical for downstream analysis. Please note:
- Ensure you provide the correct structure design parameters
- Pay attention to the mismatch parameter settings
- Monitor data quality metrics
Seeksoultools is able to extract the barcode and UMI sequences based on different Read1 structures. The pipeline processes the barcodes and filters Read1 and its corresponding Read2, and an updated fastq file is generated at the end of step 1.
Structure design and description
NOTE
The following are the basic symbols used in Read1 structure design:
- B: barcode bases
- L: linker bases
- U: umi bases
- X: other arbitrary bases used as placeholders
B: barcode bases L: linker bases U: umi bases X: other arbitrary bases used as placeholders
Taking the following two Read1 structures as examples: B8L8B8L10B8U8: 
B17U12: 
- Anchor-based misalignment design: In MM design, to increase the base balance of the linker portion during sequencing,1-4 bp shifted bases, called anchors, were added. The anchor determines the starting position of the barcode.

Workflow
Anchor determination:
For data with misaligned Read1 (produced by MM reagents), seeksoultools attempts to find the anchor sequence within the first 7 bases of the Read1 sequence to determine the start position of subsequent barcodes. If the anchor sequence is not found, the corresponding Read1 and R2 are considered invalid reads.
Barcode and linker correction:
After determining the starting position of the barcode, the corresponding sequence is extracted based on the structural design. When the extracted barcode sequence is found in the whitelist, it is considered a valid barcode and the read count with valid barcodes is recorded. When the barcode is not found in the whitelist, it is considered an invalid barcode.
TIP
During sequencing process, there is a certain probability that sequencing error occurs. With the presence of a whitelist, SeekSoulTools can perform barcode correction. When correction is enabled and an invalid barcode differs by one base (one hamming distance) from sequences in the whitelist:
- If there is only one matching sequence in the whitelist: The invalid barcode is corrected to that whitelist barcode
- If multiple matches exist in the whitelist: The invalid barcode is corrected to the sequence with the highest read support
The logic of linker correction is the same as that of barcode correction.
Adaptors and polyA sequence trimming
WARNING
If the trimmed Read2 length is less than the minimum length, the read will be considered invalid, which may result in loss of usable data.
In transcriptome, polyA tail and Adapter sequences introduced during library preparation, may appear at the end of Read2.We remove these contaminating sequences and make sure the trimmed Read2 length is greater than the minimum length for accurate alignment afterward. If the trimmed Read2 length is less than the minimum length, we consider the read to be invalid.
After the processing procedure described above, the data composition is shown in the following figure:
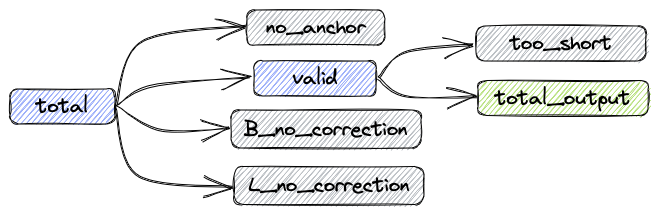
- total: Total reads
- valid: Number of reads without correction and number of successfully corrected reads
- B_corrected: Number of successfully corrected reads
- B_no_correction: Number of reads with incorrect barcodes
- L_no_correction: Number of reads with incorrect linkers
- no_anchor:Number of reads without anchors
- trimmed: Number of reads that have been trimmed
- too_short: Number of reads that are shorter than 60bps after trimming
The relationships between the metrics are as follows:
total = valid + no_anchor + B_no_correction + L_no_correction
Number of reads in the updated FASTQ file: total_output = valid - too_short
step2: Alignment
IMPORTANT
Alignment quality directly impacts downstream analysis results. Please pay special attention to:
- STAR version compatibility with reference genome index
- Alignment parameter selection
- Proportion of reads mapping to exonic and intronic regions
Sequence Alignment
- Seeksoultools uses an aligner software called STAR to perform splicing-aware alignment of reads to the genome.
- After the alignment procedure has mapped the reads to the genome, seeksoultools uses another software called qualimap along with a gene annotation transcript file GTF to bucket the reads into exonic, intronic, and intergenic regions.
- Using featureCounts, the reads aligned to the genome were annotated to genes, with different rules such as strand specificity and features used for annotation. When using exon for annotation, a read is annotated to the corresponding gene if over 50% of its bases are aligned to the exon. When using transcripts for annotation, a read is annotated to the corresponding gene if over 50% of its bases are aligned to the transcript.
NOTE
featureCounts annotation rules:
- When using exon quantification, a read is annotated to the corresponding gene if over 50% of its bases align to the exon
- When using transcript quantification, a read is annotated to the corresponding gene if over 50% of its bases align to the transcript
After the processing procedure described above, the metrics is shown below:
- Reads Mapped to Genome: Fraction of reads that aligned to the genome (including both uniquely mapped reads and multi-mapped reads)
- Reads Mapped Confidently to Genome: Fraction of reads that uniquely mapped to the genome
- Reads Mapped to Intergenic Regions:Fraction of reads that mapped to intergenic regions
- Reads Mapped to Intronic Regions:Fraction of reads that mapped to intronic regions
- Reads Mapped to Exonic Regions:Fraction of reads that mapped to exonic regions
step3: Counting
WARNING
Key considerations during the quantification process:
- UMI deduplication has a significant impact on expression estimation
- Cell calling threshold settings should be adjusted according to experimental design
- Carefully check if the number of cells matches expectations
UMI counting
Seeksoultools extracts group of reads that shared the same barcode from output BAM file and counts the number of UMIs annotated to genes and the number of reads corresponding to each UMI.
CAUTION
During UMI quantification, the following reads will be filtered out:
- Reads with UMIs consisting of repetitive bases (e.g., TTTTTTTT)
- Reads mapped to multiple genes
UMI correction
NOTE
UMIs may also have sequencing errors during sequencing process. By default, seeksoultools uses the adjacency method from UMI-tools to correct UMIs.
 Image source: https://umi-tools.readthedocs.io/en/latest/the_methods.html
Image source: https://umi-tools.readthedocs.io/en/latest/the_methods.html
Cell calling
IMPORTANT
SeekSoulTools uses the following steps to determine whether a barcode represents a cell:
- Sort all barcodes in descending order based on their UMI counts
- Calculate the threshold by dividing the UMI count at the 1% position of estimated cells by 10
- Barcodes with UMI counts above the threshold are considered cells
- Barcodes with UMI counts below the threshold but above 300 are analyzed using DropletUtils
- Barcodes that don't meet the above criteria are considered background
TIP
DropletUtils analysis method:
- Assumes barcodes with UMI counts below 100 are empty droplets
- Calculates background RNA expression profile based on total UMI counts for each gene across droplets
- Identifies cells through statistical testing of UMI counts
After the processing procedure described above, the metrics is shown below:
- Estimated Number of Cells: Total number of cells by cell calling
- Fraction Reads in Cells: Fraction of reads after cell calling among all reads used for counting
- Mean Reads per Cell: Average number of reads per cell, Total number of reads/Number of cells after cell calling
- Median Genes per Cell: Median gene count in barcodes after cell calling
- Median UMI Counts per Cell: Median UMI count in barcodes after cell calling
- Total Genes Detected: Number of genes detected in all cells
- Sequencing Saturation: 1 - Total UMI count/Total read count
step4: Downstream analysis
Seeksoultools perform downstream analysis when we have gene expression matrix from step3.
Seurat analysis
NOTE
SeekSoulTools uses Seurat to perform the following analysis steps:
- Calculate mitochondrial content
- Count total UMIs per cell
- Count total genes per cell
- Normalize the matrix
- Identify highly variable genes
- Perform dimensionality reduction
- Find differentially expressed genes
Seeksoultools use Seurat to calculate the mitochondrial content, number of genes, and UMIs of each cell. After that, the gene expression matrix is normalized, and a subset of features that exhibit high cell-to-cell variation in the dataset is identified. Linear dimensional reduction using PCA is then performed, and the result is passed to t-SNE and UMAP for visualization. A graph-based clustering procedure is then followed, and cells are partitioned into different clusters. Finally, seeksoultools finds markers that define clusters via differential expression.
